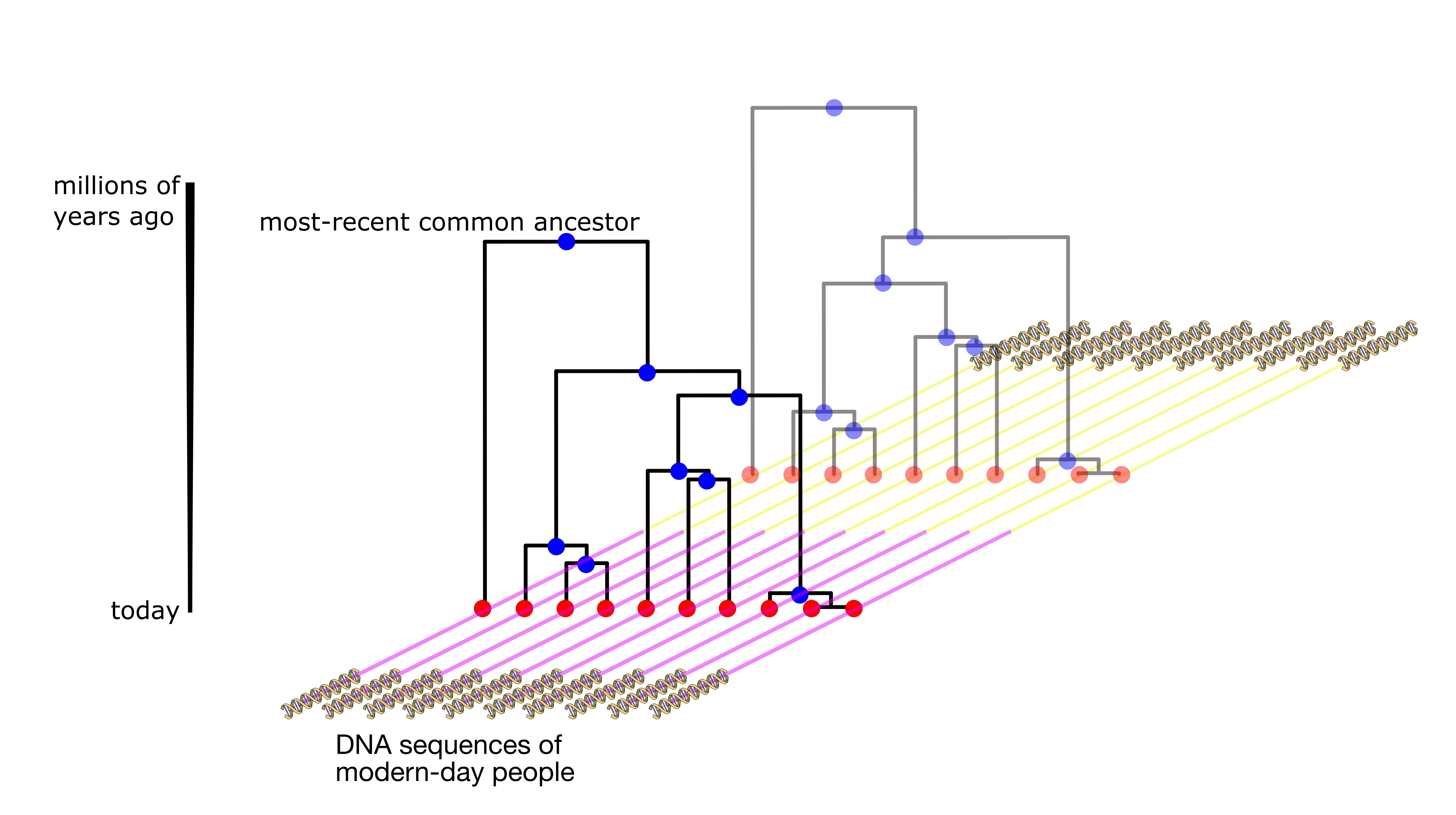
Genealogies describe the genetic relationships of individuals back in time, typically up to millions of years for humans. Relate estimates genealogies genome-wide from genetic variation data collected at the tips of these genealogies; the reconstructed trees adapt to changes in local ancestry caused by recombination. The method is scalable to thousands of samples and can be used to study population size histories, split times of populations, introgression, dating mutations, and positive selection.
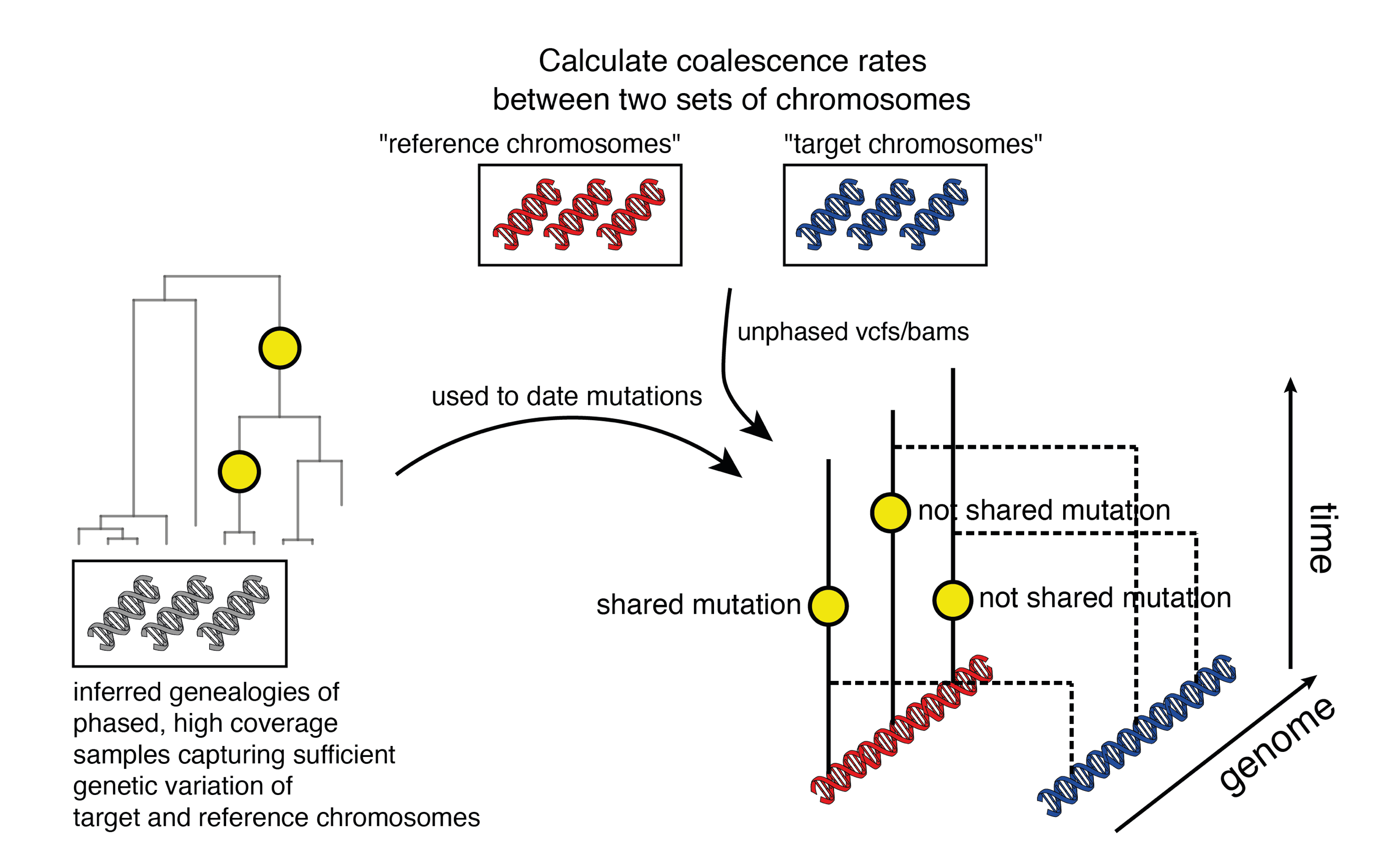
Colate leverages mutations dated using a Relate genealogy to infer coalescence rates for lower coverage genomes, specified in BCF or BAM format. These can be used infer population histories, incl recent shared ancestry, migrations/population replacements, or date population splits.

I am interested in summarising network structure in meaningful ways (e.g., by finding densely connected parts of a network), as well as understanding how the structure of a network impacts processes happening on top of them. The structure of a network representing human-human interactions is, for instance, known to influence how quickly an epidemic can spread.
We looked into how temporally evolving connectivity relationships impact such processes: diverse types of networks have non-trivial temporal patterns, which can lead to dramatic differences in e.g., epidemics spreading on such networks.